Our expertise extends to bioinformatics and/or biostatistics analyses of your data. We developed several automated analysis pipelines, based on the best available algorithms. We have dozens of calculation servers in house in order to run our pipelines, together with sufficient storage space.
The amounts and quality of data produced by next generation sequencers evolve regularly. Our laboratory adapts continuously and proposes tools to answer your questions and get the best out of your data: mapping and variant discovery with exome and genome sequencing, annotation of discovered variants using public data (Ensembl, Encode, …), family and epigenetics studies such as transcriptome and methylome analyzes and protein-DNA interactions. We also provide various statistics assuring quality control of the produced data.
The Biostatistics group is composed of 4 biostatisticians. They take care of :
Expertise
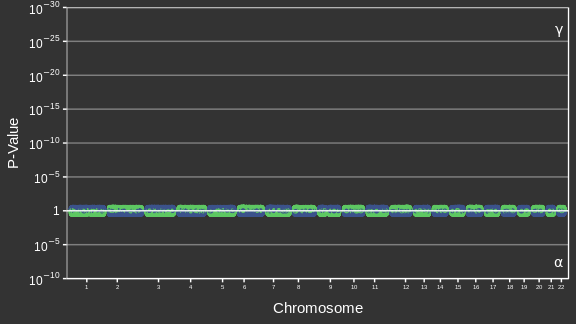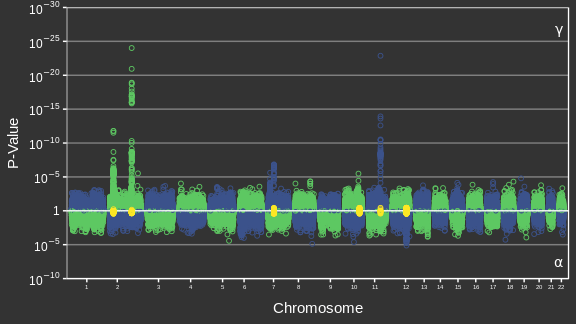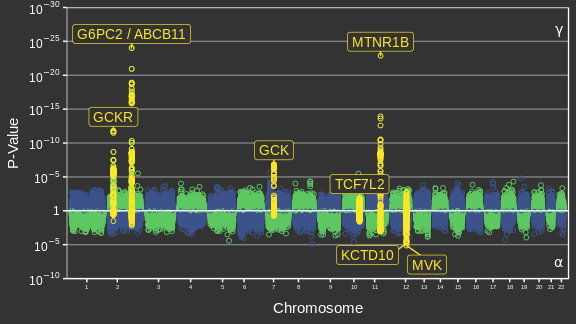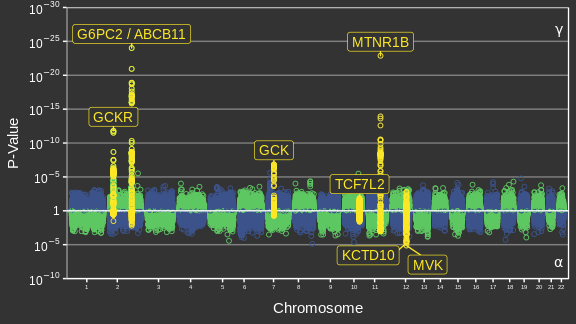
Adaptée de Canouil et al. (2018)
Pipelines & Extension R (https://github.com/umr1283/)
“Detection of Clonal Mosaic Events” : http://www.good.cnrs.fr/app/uploads/2012/09/CALLME.zip
snpEnrichment : https://CRAN.R-project.org/package=snpEnrichment
clere : https://CRAN.R-project.org/package=clere
NACHO : https://m.canouil.fr/NACHO/, https://CRAN.R-project.org/package=NACHO
insane : https://m.canouil.fr/insane/, https://CRAN.R-project.org/package=insane
The Bioinformatics group is composed of 4 bioinformaticians. They take care of :
Main future development projects are :

