We are able to deliver you extracted samples, high throughput genotyping data, high throughput sequencing data, biostatistics and NGS bioinformatics analyses. We are Illumina Service Provider (CSPro) for sequencing on NGS equipments.
Extractions
Jellyfish of DNA in suspension © F.Viala/CNRS Photothèque
Genotyping, expression and kinome

Illumina chips for genotyping or expression studies.
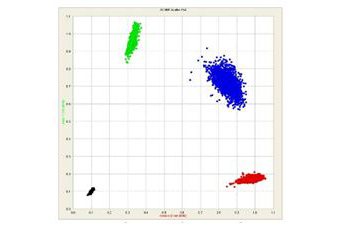
SNP genotyping scatter plot : genotype distribution as clusters after analysis
NGS Sequencing
Libraries preparation for High throughput sequencing:
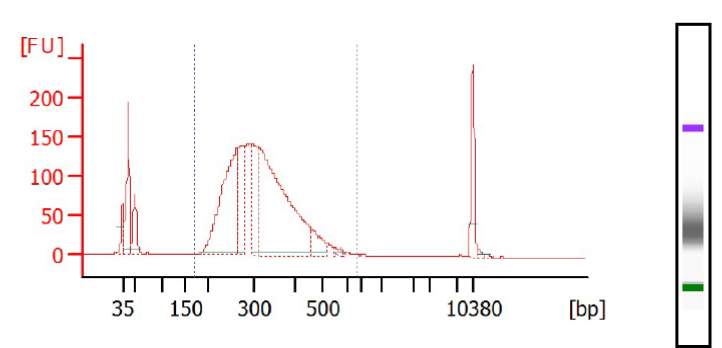
Profiles of Whole Exome (KAPA HyperCap With HyperExome Probes) obtained via Bioanalyzer, DNA High Sens chip – These libraries were fragmented and prepared via Sciclone® G3 NGSx iQ ™ Workstation (PerkinElmer®). They are ready for targeted sequencing via our Illumina sequencers (NovaSeq or NextSeq).
Next Generation Sequencing (NGS) using Illumina NovaSeq6000, NextSeq500, MiSeq
We acquired the NovaSeq in June 2017, the first equipment of this type installed in France at the time! The NextSeq500 was acquired in the first half of 2015. The MiSeq dates from the end of 2013.
These equipments allow us to offer you three sequencing modes: the ultra high, the high and the medium throughput sequencing modes. Whatever the method chosen, any types of libraries are susceptible to be sequenced on the HiSeq: Exomes, small RNA, total RNA, mRNA, MethylSeq, ChipSeq, amplicons, genomes, RAD…
We are certified for sequencing on Illumina NGS equipments.

The Sequencing Analysis Viewer tool (Illumina) allows the visualization of the sequencing quality parameters during a run or at the end of a run on our sequencers NovaSeq, NextSeq or MiSeq.
A 1: flow cell chart displays color-coded quality metrics per tile for the entire flow cell (4 lanes of a NovaSeq S4 run); 2: Cycle plot displays plots that allows to follow the progression of quality metrics during a run, each base is color-coded; 3: QScore distribution: allows to visualize the number of reads by quality score. The quality score is cumulative for current cycle and previous cycles and only reads that pass the quality filter are included; 4: data per lane (numbdf of clusters/mm²) ; 5: QScore Heatmap : displays plots that allows to view the QScore by cycle. B Imaging tab: displays in detail a section of a flow cell. Various quality parameters can be analyzed (density of clusters pass clusters, error rate (determined by alignment with PhiX)…).
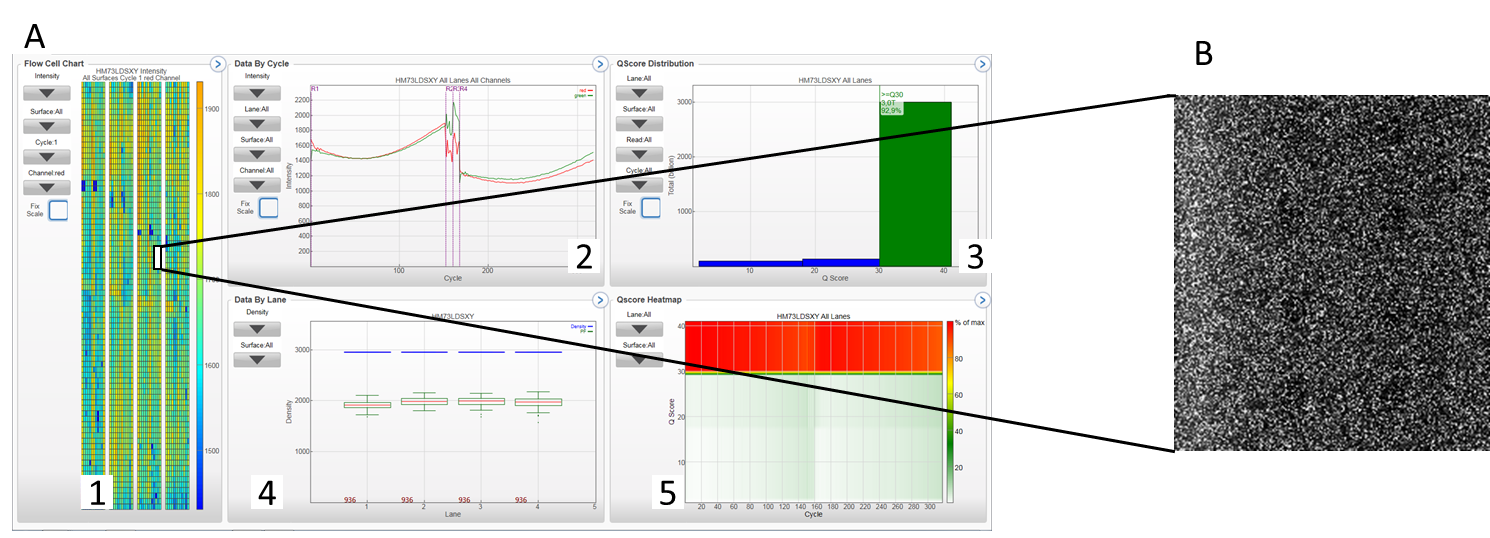
Overview of a sequencing run on NovaSeq / Image of a section of a flow cell
Bioinformatic and bio-statistics analyzes
Bioinformatics analysis of high-throughput sequencing data :
Whole exome / Whole genome :
We offer different levels of analysis :

Sequence alignment from high throughput sequencers after use of Integrative Genomics Viewer tool